library(irr)Loading required package: lpSolveWarning: package 'lpSolve' was built under R version 4.3.3library(yardstick)
Attaching package: 'yardstick'The following object is masked from 'package:readr':
speclibrary(irr)Loading required package: lpSolveWarning: package 'lpSolve' was built under R version 4.3.3library(yardstick)
Attaching package: 'yardstick'The following object is masked from 'package:readr':
spec# Diagnoses data
data("diagnoses", package = "irr")
diagnoses rater1 rater2 rater3
1 4. Neurosis 4. Neurosis 4. Neurosis
2 2. Personality Disorder 2. Personality Disorder 2. Personality Disorder
3 2. Personality Disorder 3. Schizophrenia 3. Schizophrenia
4 5. Other 5. Other 5. Other
5 2. Personality Disorder 2. Personality Disorder 2. Personality Disorder
6 1. Depression 1. Depression 3. Schizophrenia
7 3. Schizophrenia 3. Schizophrenia 3. Schizophrenia
8 1. Depression 1. Depression 3. Schizophrenia
9 1. Depression 1. Depression 4. Neurosis
10 5. Other 5. Other 5. Other
11 1. Depression 4. Neurosis 4. Neurosis
12 1. Depression 2. Personality Disorder 4. Neurosis
13 2. Personality Disorder 2. Personality Disorder 2. Personality Disorder
14 1. Depression 4. Neurosis 4. Neurosis
15 2. Personality Disorder 2. Personality Disorder 4. Neurosis
16 3. Schizophrenia 3. Schizophrenia 3. Schizophrenia
17 1. Depression 1. Depression 1. Depression
18 1. Depression 1. Depression 1. Depression
19 2. Personality Disorder 2. Personality Disorder 4. Neurosis
20 1. Depression 3. Schizophrenia 3. Schizophrenia
21 5. Other 5. Other 5. Other
22 2. Personality Disorder 4. Neurosis 4. Neurosis
23 2. Personality Disorder 2. Personality Disorder 4. Neurosis
24 1. Depression 1. Depression 4. Neurosis
25 1. Depression 4. Neurosis 4. Neurosis
26 2. Personality Disorder 2. Personality Disorder 2. Personality Disorder
27 1. Depression 1. Depression 1. Depression
28 2. Personality Disorder 2. Personality Disorder 4. Neurosis
29 1. Depression 3. Schizophrenia 3. Schizophrenia
30 5. Other 5. Other 5. Other
rater4 rater5 rater6
1 4. Neurosis 4. Neurosis 4. Neurosis
2 5. Other 5. Other 5. Other
3 3. Schizophrenia 3. Schizophrenia 5. Other
4 5. Other 5. Other 5. Other
5 4. Neurosis 4. Neurosis 4. Neurosis
6 3. Schizophrenia 3. Schizophrenia 3. Schizophrenia
7 3. Schizophrenia 5. Other 5. Other
8 3. Schizophrenia 3. Schizophrenia 4. Neurosis
9 4. Neurosis 4. Neurosis 4. Neurosis
10 5. Other 5. Other 5. Other
11 4. Neurosis 4. Neurosis 4. Neurosis
12 4. Neurosis 4. Neurosis 4. Neurosis
13 3. Schizophrenia 3. Schizophrenia 3. Schizophrenia
14 4. Neurosis 4. Neurosis 4. Neurosis
15 4. Neurosis 4. Neurosis 5. Other
16 3. Schizophrenia 3. Schizophrenia 5. Other
17 4. Neurosis 5. Other 5. Other
18 1. Depression 1. Depression 2. Personality Disorder
19 4. Neurosis 4. Neurosis 4. Neurosis
20 5. Other 5. Other 5. Other
21 5. Other 5. Other 5. Other
22 4. Neurosis 4. Neurosis 4. Neurosis
23 5. Other 5. Other 5. Other
24 4. Neurosis 4. Neurosis 4. Neurosis
25 4. Neurosis 4. Neurosis 5. Other
26 2. Personality Disorder 2. Personality Disorder 4. Neurosis
27 1. Depression 5. Other 5. Other
28 4. Neurosis 4. Neurosis 4. Neurosis
29 3. Schizophrenia 3. Schizophrenia 3. Schizophrenia
30 5. Other 5. Other 5. Othercm_r1r2 <- conf_mat(diagnoses, truth = rater1, estimate = rater2)
autoplot(cm_r1r2, type = "heatmap")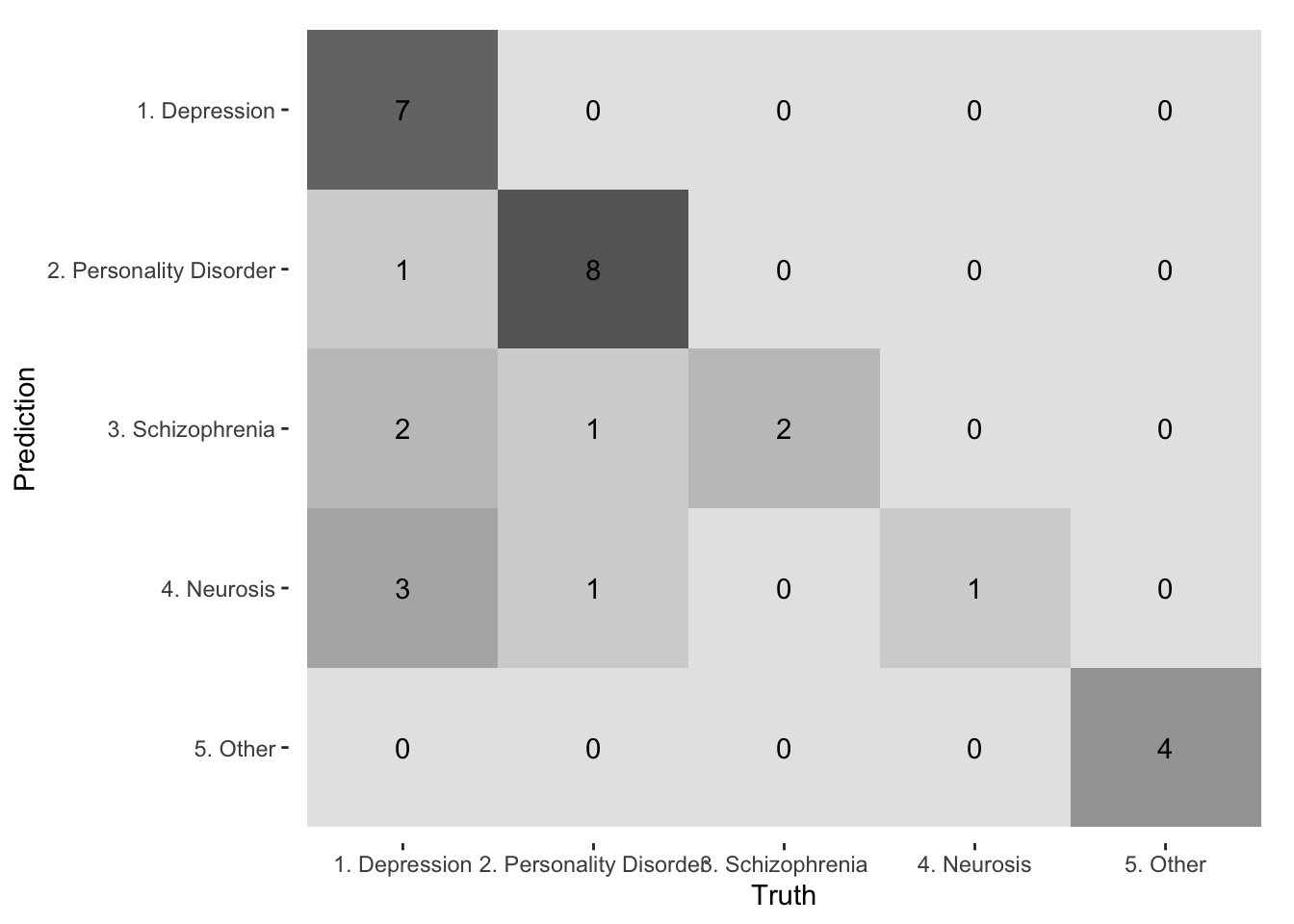
The Cohen’s kappa corresponds to the unweighted kappa. It can be used for two nominal or two ordinal categorical variables
diagnoses |>
select(rater1, rater2) |>
kappa2(weight = "unweighted") Cohen's Kappa for 2 Raters (Weights: unweighted)
Subjects = 30
Raters = 2
Kappa = 0.651
z = 7
p-value = 2.63e-12 data("anxiety", package = "irr")
head(anxiety, 4) rater1 rater2 rater3
1 3 3 2
2 3 6 1
3 3 4 4
4 4 6 4icc(
anxiety, model = "twoway",
type = "agreement", unit = "single"
) Single Score Intraclass Correlation
Model: twoway
Type : agreement
Subjects = 20
Raters = 3
ICC(A,1) = 0.198
F-Test, H0: r0 = 0 ; H1: r0 > 0
F(19,39.7) = 1.83 , p = 0.0543
95%-Confidence Interval for ICC Population Values:
-0.039 < ICC < 0.494# Simulated radiomics features data for 5 subjects and 2 observers
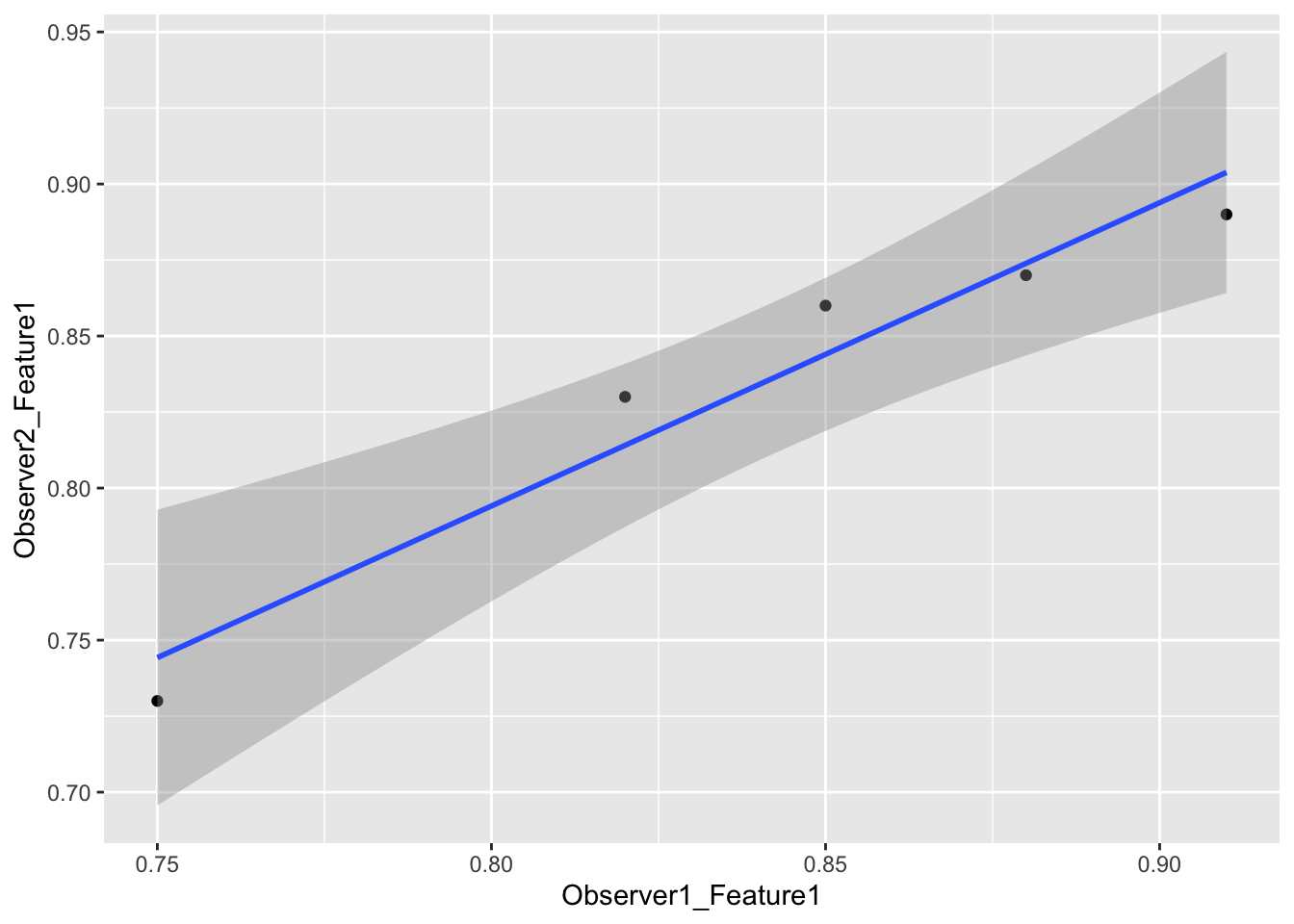
radiomics_data <- data.frame(
Subject = 1:5,
Observer1_Feature1 = c(0.75, 0.82, 0.91, 0.85, 0.88),
Observer2_Feature1 = c(0.73, 0.83, 0.89, 0.86, 0.87),
Observer1_Feature2 = c(100, 102, 105, 108, 107),
Observer2_Feature2 = c(99, 101, 104, 109, 106)
)
radiomics_data Subject Observer1_Feature1 Observer2_Feature1 Observer1_Feature2
1 1 0.75 0.73 100
2 2 0.82 0.83 102
3 3 0.91 0.89 105
4 4 0.85 0.86 108
5 5 0.88 0.87 107
Observer2_Feature2
1 99
2 101
3 104
4 109
5 106# ICC for Feature 1 across both observers
feature1_data <- radiomics_data[, c("Observer1_Feature1", "Observer2_Feature1")]
icc_feature1 <- icc(feature1_data,
model = "twoway",
type = "agreement",
unit = "single")
icc_feature1 Single Score Intraclass Correlation
Model: twoway
Type : agreement
Subjects = 5
Raters = 2
ICC(A,1) = 0.972
F-Test, H0: r0 = 0 ; H1: r0 > 0
F(4,4.95) = 66.4 , p = 0.000171
95%-Confidence Interval for ICC Population Values:
0.805 < ICC < 0.997radiomics_data |>
ggplot(aes(Observer1_Feature1, Observer2_Feature1)) +
geom_point() +
geom_smooth(method = "lm")`geom_smooth()` using formula = 'y ~ x'